The Phytomine Cookbook
How to do common operations in Phytomine
The InterMine interface of Phytozome allows anyone to conduct powerful queries over the entire dataset in Phytozome. Doing so may require a little trial and error at first in order to understand the structure of the data, but after mastering the structure very powerful analyses can be performed on the data. This guide provides some examples that can be the starting point for your queries.
There are also some video tutorials produced from FlyMine on using InterMine. Additionally, other web sites using InterMine, such as WormMine and YeastMine have online help documents and screencasts which illustrate the use of the interface. While the contents of the databases are different, the structure of the web pages is the same.
Lists
Many queries start or end with making a list of identifiers - such as names of genes, proteins, protein families, and so on. The easiest way to create a list from identifiers you already know is to click on the Lists link on the top tab bar and simply type, paste or upload a file of your identifiers. Then press 'Create List'. The identifiers must be of one type; you cannot put genes and protein identifiers into a single list.
After hitting the create list button, any identifiers not found or duplicated in your list are reported. You may want to check those. If all look correct, give the list a name you can recognize and save the list.
Login
We recommend that you create an account with us so that you can save your lists on our server. This will allow you to keep your results and not have to upload them repeatedly. Accounts are free for all users and do not require any approvals. You will use the same login account if you want to do a bulk download of data from our genome portal
Basic Queries
| Front Page |
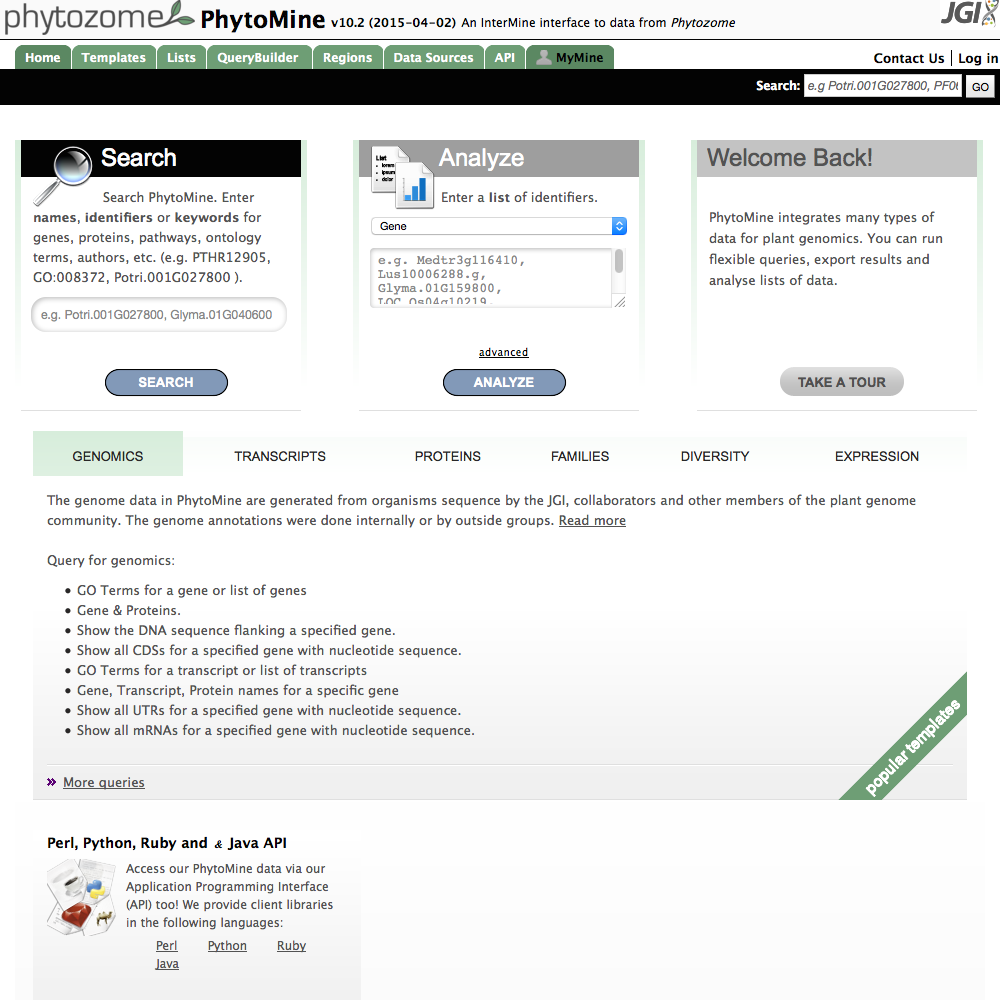 |
| Click image to enlarge. |
Many basic queries have templates which you can access from the front page. They are grouped into different aspects of the data. Selecting the different tabs - Genomics, Transcripts, Proteins,... - allows you to view the types of queries available.
| A Simple Template |
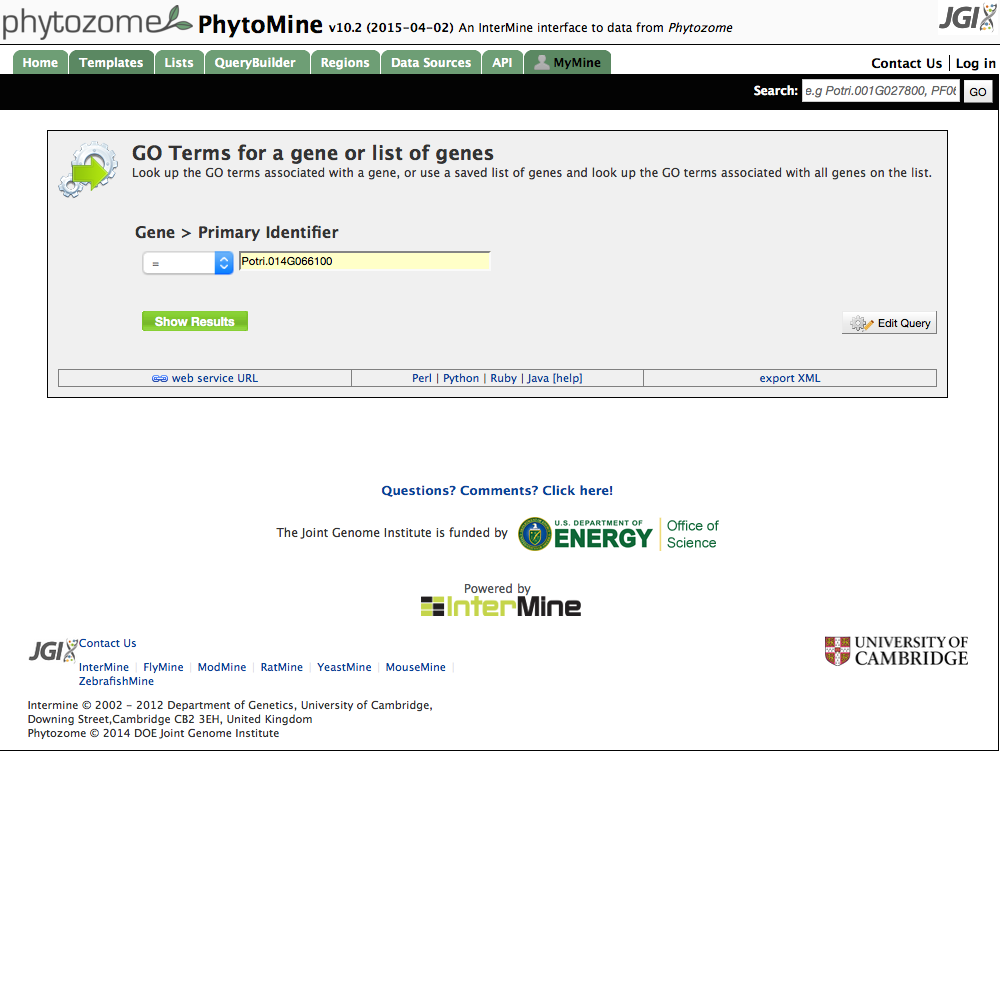 |
| Click image to enlarge. |
These are simple examples of common types of queries you can make. There are examples of finding all proteins for a specific gene, or all proteins in an organism and can be used to generate large tables of data for downloading. Some of them (for example, all proteins with a specific PFAM domain) are very specific and generate only a few results. By clicking on one of the templates, then the 'Edit Query' button, the templated query may be modified to formulate a query that is customized for your needs.
| All Query Templates |
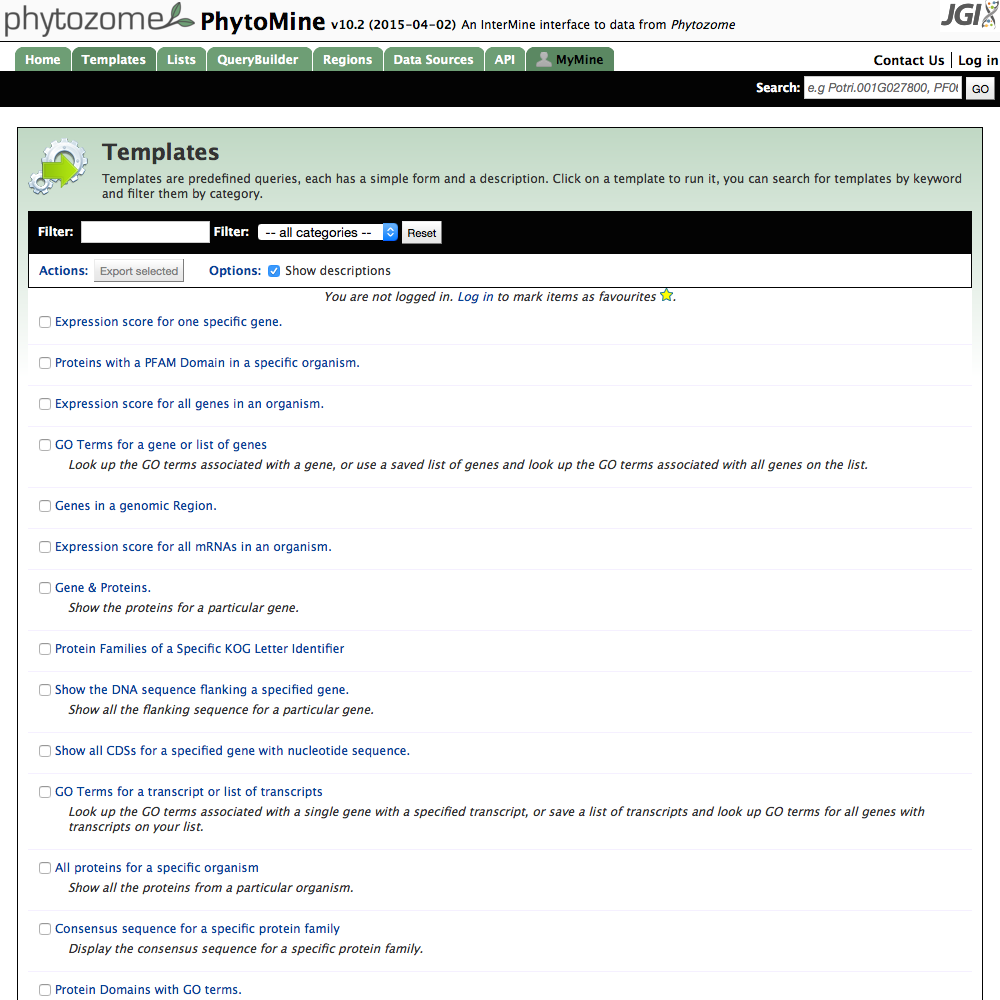 |
| Click image to enlarge. |
Additionally there is a longer list of templated queries accessible from the Templates tab on the home page menubar. This page shows a list lf all templates available on the web site. More can be added, and if there you find a need for a particular one which you think would be useful to the community, please contact us so that we can make it available.
Custom Queries
| 1 Start a Query |
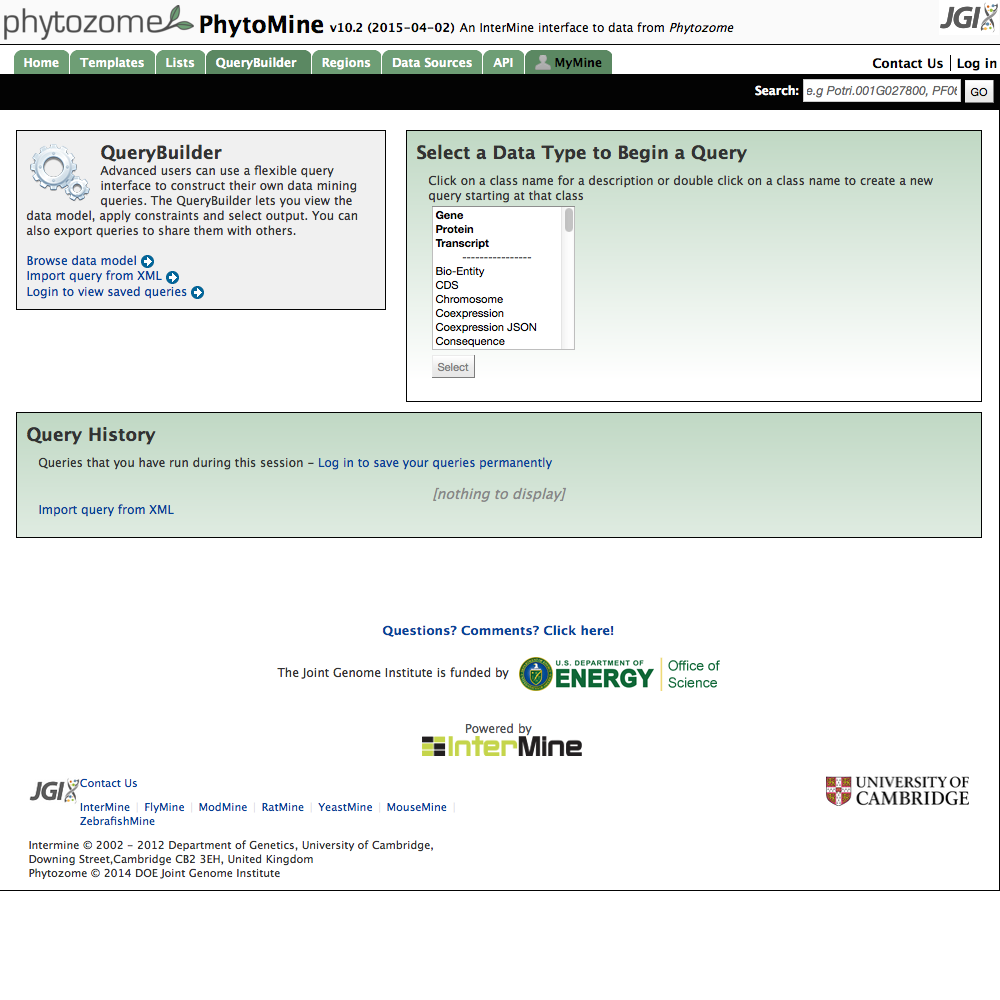 |
| Click image to enlarge. |
For your own query, start by going to the Query Builder page.
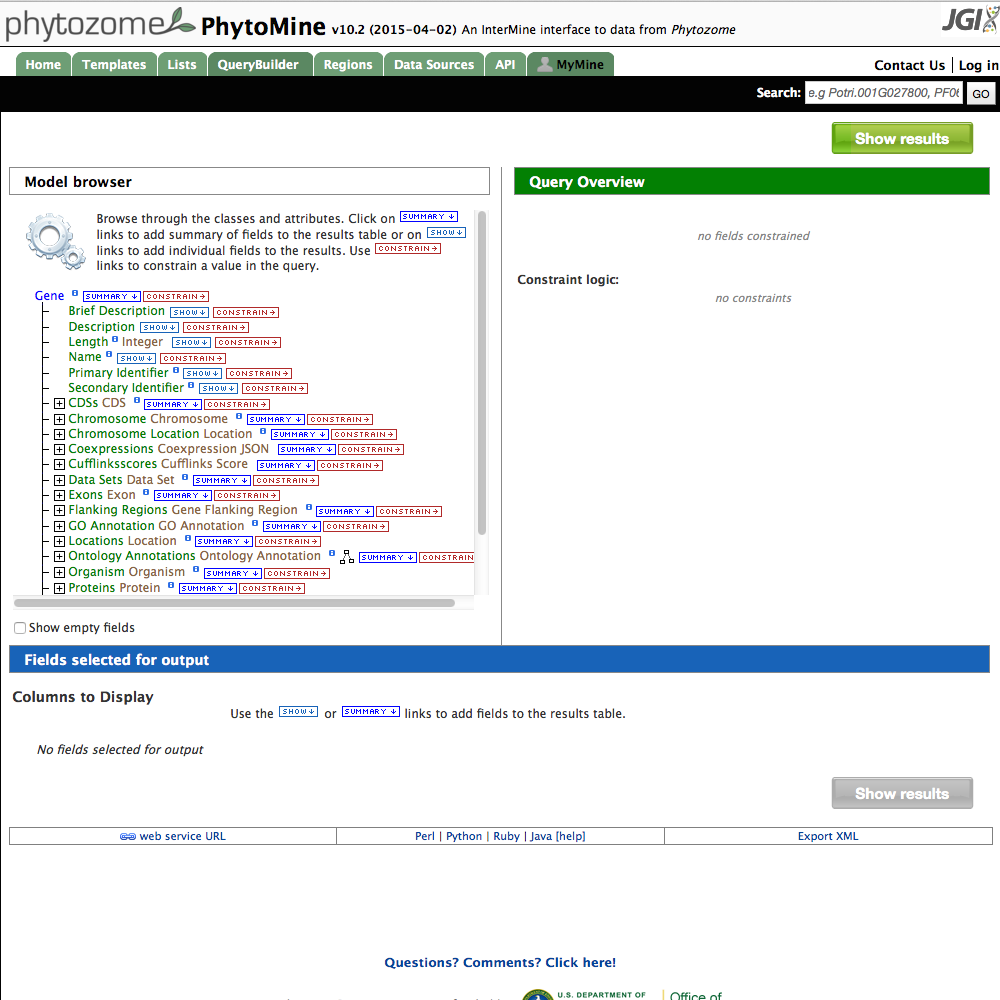
| 2 Select Gene |
 |
| Click image to enlarge. |
- Start by selecting the starting data type for the query. This is often 'Gene' or 'Protein'. Click on the 'Gene' label in the right column, then hit the 'Select' button below it.
- In the Model Browser pane, you can see the fields available for selection. Click on the 'SHOW' button next to 'DB identifier'. This will include the gene name in the results field.
| 3 Expand Protein |
 |
| Click image to enlarge. |
- Each square with a small + sign is a data item that is linked to the gene. If you want to find the proteins linked to a gene, click the box next to 'Proteins'.
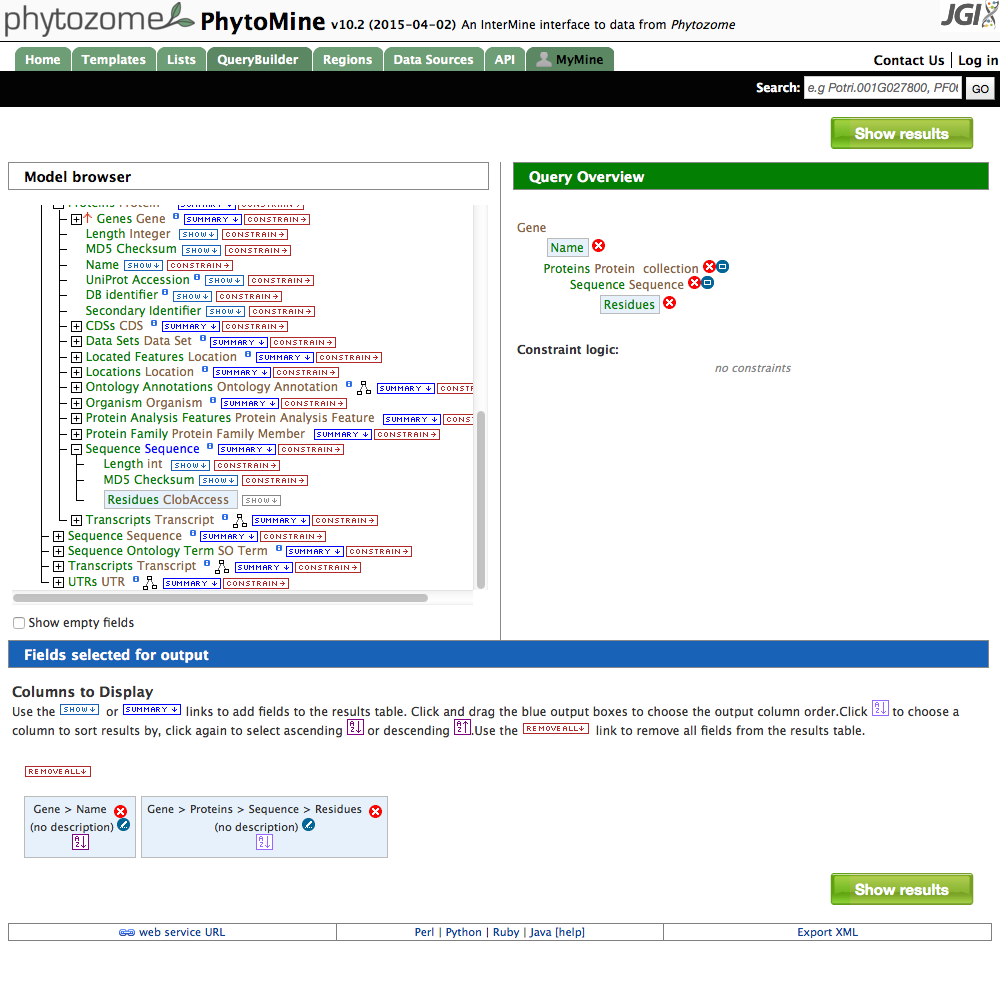
| 5 Select Residues |
 |
| Click image to enlarge. |
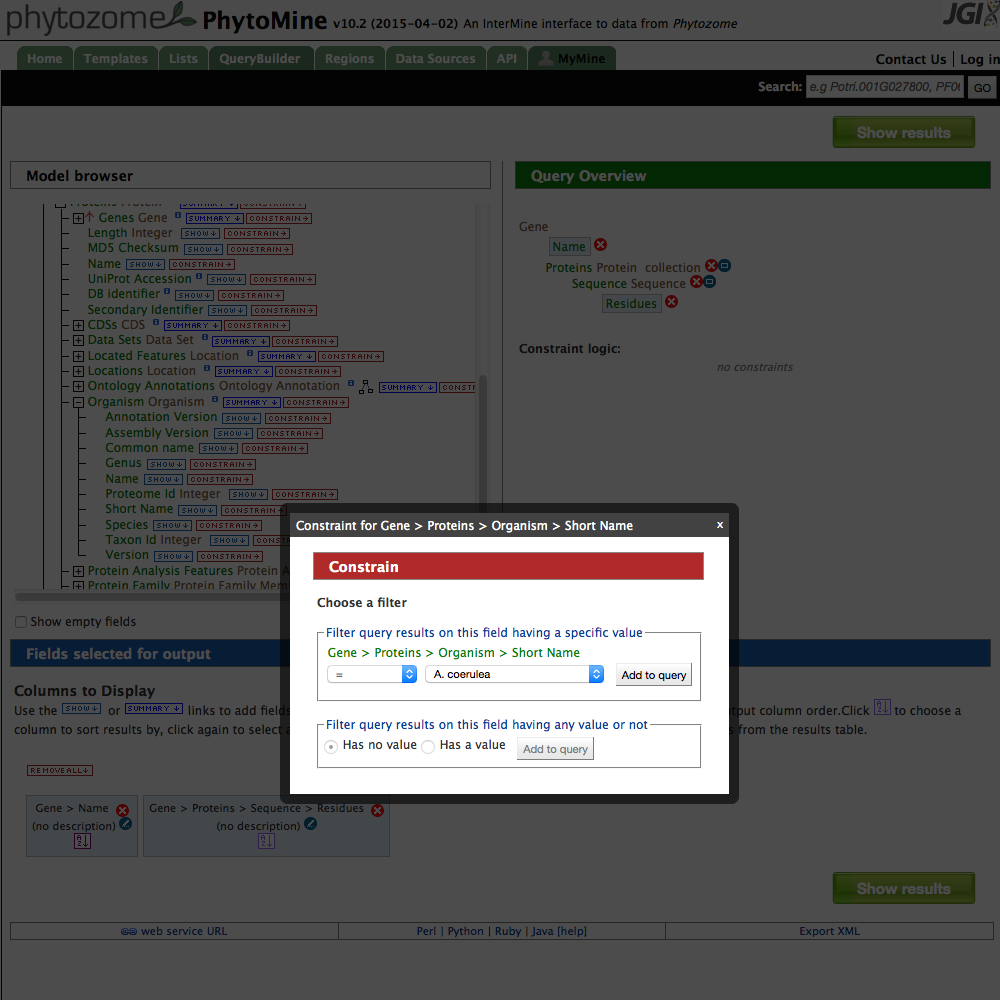
| 6 Constrain Organism |
 |
| Click image to enlarge. |
- With the Proteins box expanded (notice the + sign is now a - sign), you can select attributes of the protein. Click the 'SHOW' box next to 'DB identifier' to include the protein name in the results.
- Now expand the 'Sequence' box that is connected to the 'Proteins' label. This is the sequence information attached to the protein. Click on the 'SHOW' box next to 'Residues'.
- Finally, rather than querying for every protein sequence, restrict the search to a particular organism. Click on the + sign next to 'Organism'. Click on the 'CONSTRAIN' button next to 'Short Name'. A dialog box open and you can select one of the organisms in phytomine. Select one and then press 'Add to query'.
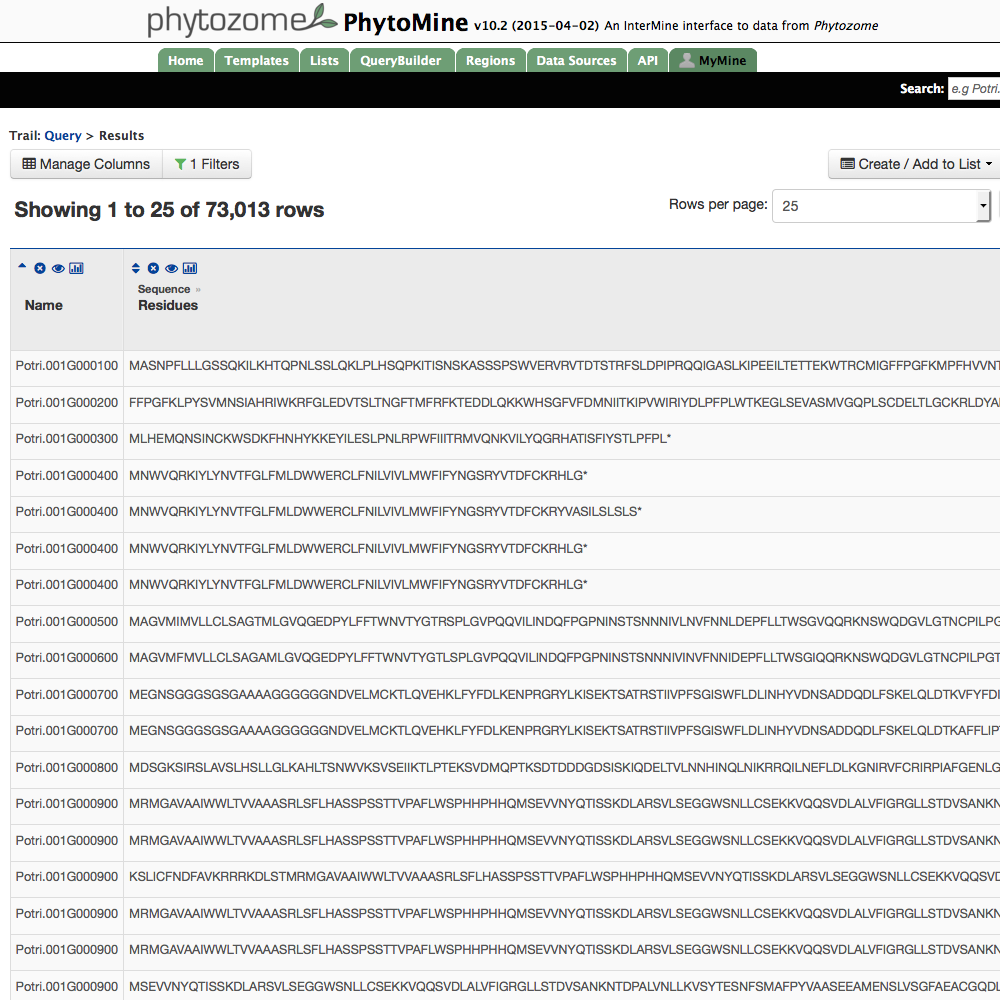
| View Results |
 |
| Click image to enlarge. |
The query is now ready to run. Press 'Show results' and the output will appear in a few seconds (or more, depending on the complexity of the query and the number of results).
If you had created a list of gene names previously and now want to find the proteins associated with them then do this.
- From the results page, click on the 'Query' link on the left side of the results page above the table of results. This will bring you back to query builder page.
- Click on the small red X next to the name of the organism you have selected in the previous query. This eliminates the organism constraint you previously used.
- Click on the 'CONSTRAIN' button next to Gene->DB identifier. A dialog box will open and select the check box next the text 'Contained in list:' and select the list you have created. This option will only be present if you have already created a list. Then select 'Add to query'.
- Again, press the 'Show results' button to retrieve the proteins for your list of genes.
The results of a query can be used to create a new list. Suppose you want to create a list of proteins based on this query.
- From the results page, select the pulldown menu labeled 'List' on the right side above the table of results.
- Select 'Create New List" and 'All Proteins'.
- You will see a dialog box that will allow you to give a name, description and optional category tags to this list.
Advanced Queries
The gene and protein queries are a good starting point for basic annotation queries. Other, more complex queries, will typically involve other starting points.
- Computed features of the protein sequence start with the 'Protein Analysis Feature' table. From this starting point, you can select results from any of the InterproScan results. If you are interested in a specific PFAM domain, constrain the 'Cross Reference'->'Identifier' to the one you are interested in, and constrain the 'Organism'->'Short Name' to the one you are studying. Be sure to select 'Protein'->'DB identifier' to output the name of the protein.
- Starting with 'Protein Family' will give you clusters of similar proteins. Constraining 'Protein'->'DB identifier' will search for a particular protein by name. Including 'Sequence Alignment'->'MSA' will show you the multiple sequence alignment of that protein in each cluster it is a member of. Or, 'Centroid Sequence'->'Residues' will show you a consensus sequence of that cluster.
- Nucleotide variation in natural populations is available in the 'SNP' table. To restrict the search to a particular organism (Note that we do not have data on all organisms present in Phytozome), select 'SNP'->'Snp Diversity Samples'->'Diversity Sample'->'Organism'. From the 'Consequences' link you can restrict you search to find SNPs that affect only particular genes.
- RNA-Seq expression data are available from the starting point 'Cufflinks Score'. The results are only available for a few organisms.
Saving Queries
As mentioned previously, one column of results from a query can be saved as a list for future use. You can also combine multiple lists by merging, looking for intersections or differences.
You may find it easiest to export the XML for a particular query and save it locally. To do this, select the 'Code'->'XML' from the dropdown menu on the results table. If you examine this XML you can see how the 'view' and 'constraint' fields of the XML match what you have selected from the graphical interface. Experiment by editing the XML by adding or removing fields from the view to generate more results if you desire. You can paste the XML at some later by selecting the 'Import Query from XML' on the Query Builder page. Be careful that while copying, pasting or editing the XML, your software does not convert any of the double-quote marks (") to either open-double-quotes (“) or closed-double-quotes (”). If these latter symbols are used, the query will generate a cryptic error message.
If a query is likely to generate a very large number of rows, the query planner will not execute the request in order to prevent too much load on our servers. If you have a query which is not returning any results, please try to constrain your query to reduce the number of results and try again.
Downloading Results
From the results page, you can save the results of a query as a list. If you have logged into our web portal, this list is maintained in our server for your reuse later. Or, you can download to a file on your computer by selecting the 'Download' button in the upper right corner of the results page. Results can be saved in comma or tab separated file for use with spreadsheets, XML or JSON for use with computation parsing programs, GFF3 or UCSC-BED for use with genome browsers, or raw FASTA sequence. The choice of format partly depends on the nature of the results since not all output formats are suitable for some queries. For example, you cannot save a GO term lookup in GFF3 format.
When saving a FASTA file, the downloaded results may differ from the view of the results on the web page. It is not necessary to include the sequence in list of included columns when generating the query. If you have created a query that displays multiple columns, for example, gene name, transcript name and protein name, the created FASTA file will have only the sequence of the first column in the list. In the previous example, it would be only the gene flanking sequence. No other information will be present in the file. In this example, if you wanted to download protein sequence you must either make the protein name be the first column displayed when generating the query, or edit the column list after getting results to make it first. button on the right side of the page above the table. Just select the format you want to have the results in.
Also, if you are interested in downloads of large datasets, for example, the complete genome assembly or an organism, consider using our bulk download portal.
Contact us if you need assistance in generating a query or interpreting the results.